Cloud MR: An Open-Source Software Framework to Democratize MRI Training and Research
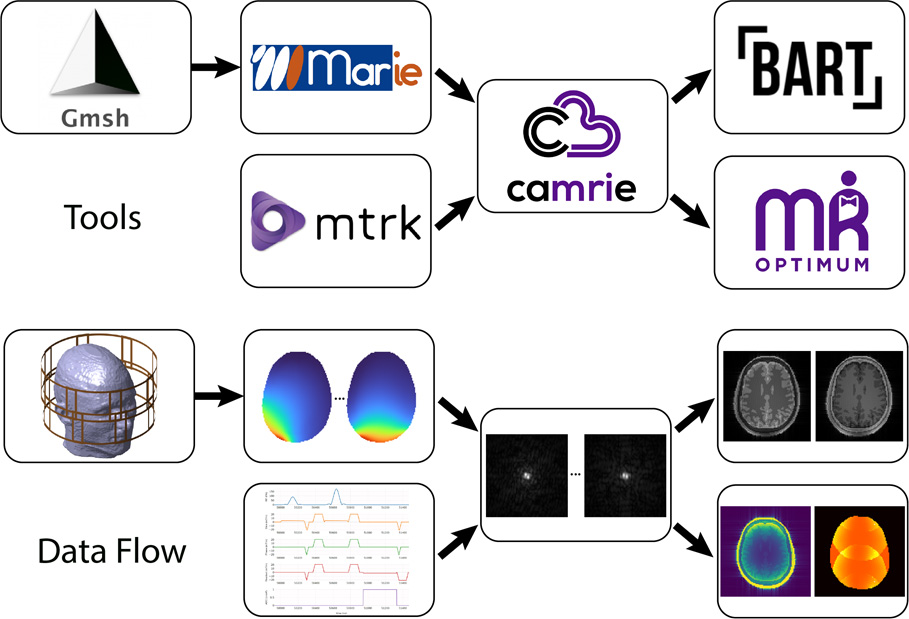
Overview
In this project we are developing an openly shared “virtual scanner” in order to broaden access to MRI experiments by decoupling them from MRI machines.
The advancement of magnetic resonance imaging technologies has had a tremendous impact on healthcare in the last half century. However, most MRI research continues to be predicated on access to scanners, an overwhelming majority of which are primarily used for clinical imaging. The resulting bottleneck has contributed to limiting research on MRI to a relative niche. The goal of this project is to relax this constraint and accelerate the pace of innovation by making available Cloud MR, an open-source simulation platform that puts a virtual scanner at the fingertips of anyone with an internet-connected web browser.
Through a web-based graphical user interface, Cloud MR aims at allowing researchers to test new MRI technologies and enabling radiologists to optimize clinical protocols without operating an MR scanner. The platform is expected to make possible radiofrequency coil modeling, pulse sequence design, and the development of image reconstruction methods—functionalities that we also anticipate to be valuable tools in education and training. Other potential benefits of simulated MRI experiments include the capacity to generate realistic synthetic MR data for training neural networks—hence to address a major research need without raising privacy concerns associated with patient data—and a decrease in MR-research-related carbon emissions.
We are developing Cloud MR with the goal of making all its constituent applications free and accessible via the portal cloudmrhub.com. Planned operating modes include the alternative to set up simulation jobs in a web browser and run them in the cloud or to download containers and run jobs on a local workstation. The source code and compiled executables are being released through GitHub at github.com/cloudmrhub.
Keywords
- MRI
- Open-Source Software
- Simulation
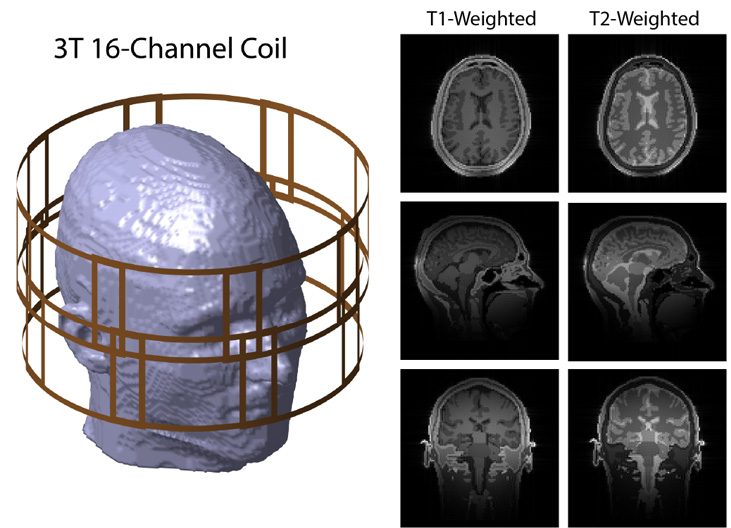
Figure 1. State-of-the-art tools were interconnected to run a comprehensive MRI simulation. Magnitude brain images for axial (top), sagittal (middle), and coronal (bottom) slices, virtually acquired with a 3 tesla 16-channel head coil. The head array was designed with gmsh, the coils’ electromagnetic fields were simulated using MARIE, the spin echo sequence was developed using mtrk, and the synthetic k-space was generated using CAMRIE. The images of the individual coils in the arrays were combined using Root Sum of Squares (RSS).
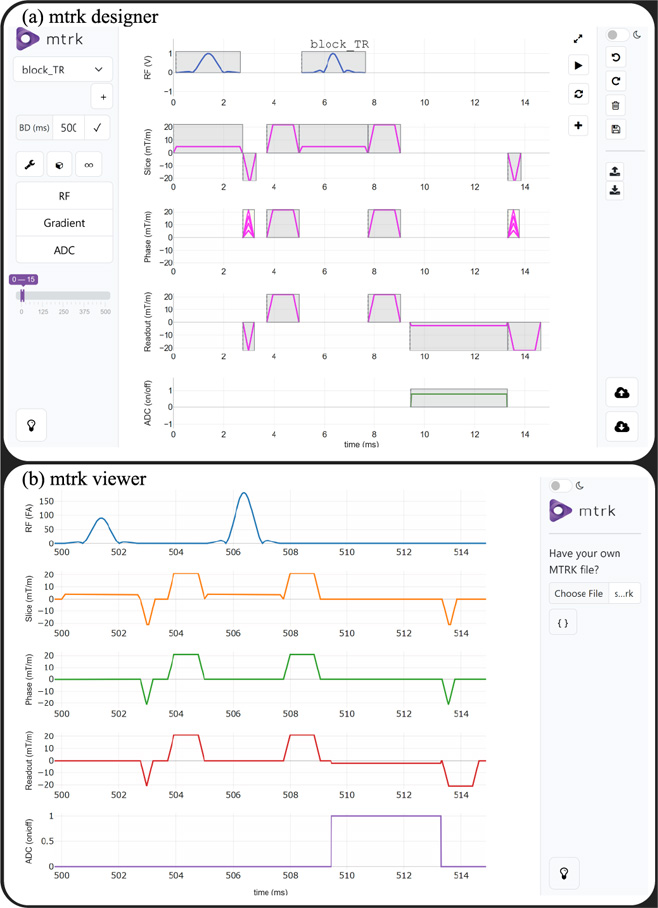
Figure 2. mtrk is a flexible open-source MRI pulse sequence development framework that enables the design of pulse sequences using a web-based graphical user interface and makes sequence development more accessible for researchers without programming experience. The mtrk designer user interface provides an intuitive graphical approach for pulse sequence development (a). Diagrams of sequences developed in mtrk or Pulseq can be displayed using the mtrk viewer (b). Note that the RF pulses are scaled based on the associated flip angle.
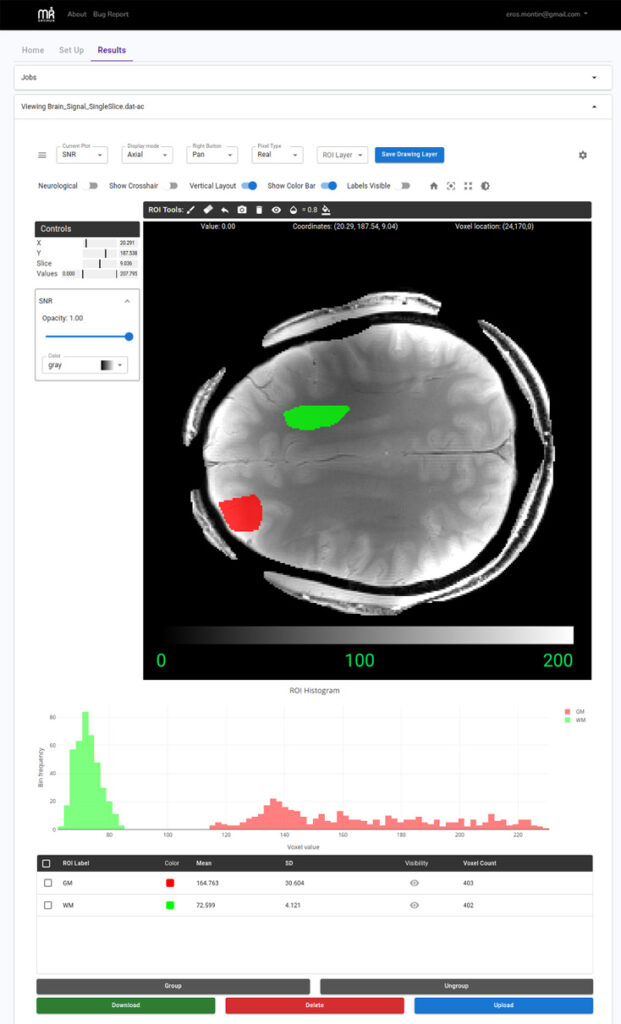
Figure 3. MR Optimum (pronounced “Mister Optimum”) is a web-based cloud-native, open-source platform for standardized signal-to-noise ratio (SNR) analysis. MR Optimum integrates established SNR estimation techniques—including multiple replicas, pseudo multiple replicas, generalized pseudo multiple replicas, and analytic methods—within a flexible, modular software architecture. The figure shows the SNR map for a representative axial slice in the brain, highlighting regional differences between a frontal area—closer to the receive coils—and a more central region. Real-time statistics and histograms of selected regions of interests (ROIs) are displayed below the map, providing an intuitive assessment of image quality.
Project Team

Riccardo Lattanzi, PhD
Project Lead
External Collaborators
- Daniele Panozzo, PhD , NYU Courant Institute of Mathematical Sciences
- Denis Zorin, PhD, NYU Courant Institute of Mathematical Sciences
- Daniel Zint, PhD, NYU Courant Institute of Mathematical Sciences
Publications
- Artiges A, Singh Saimbhi A, Lattanzi R and Block KT, mtrk – A flexible open-source framework for developing MRI pulse sequences based on common web standards; 32nd Scientific Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). Singapore, 04-09 May 2024, p. 4671.
- Serralles J, Giannakopoulos I and Lattanzi R, On the Extension of MARIE Coil Simulation to Low Frequencies and Arbitrarily Fine Meshes; 32nd Scientific Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). Singapore, 04-09 May 2024, p. 2843.
- Montin E and Lattanzi R, Rado – A Cloud-Based Toolbox for Radiomics Analysis; 31st Scientific Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). Toronto (Canada), 03-08 June 2023, p. 2889. (digital poster)
- Montin E, Carluccio G, Collins C and Lattanzi R, A web-accessible tool for temperature estimation from SAR simulations (TESS) ; 30th Scientific Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). London (UK), 07-12 May 2022, p. 582.
- Montin E, Georgakis IP, Zhang B and Lattanzi R, A software tool to assess radiofrequency coil designs with respect to ultimate intrinsic performance limits ; 30th Scientific Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). London (UK), 07-12 May 2022, p. 2780.
- Montin E, Carluccio G and Lattanzi R, A web-accessible tool for rapid analytical simulations of MR coils via cloud computing ; 29th Scientific Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). Virtual Conference, 15-20 May 2021, p. 3756.
- Montin E, Carluccio G, Collins C and Lattanzi R, CAMRIE – Cloud-Accessible MRI Emulator ; 28th Scientific Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). Virtual Conference, 08-14 August 2020, p. 1037.
- Montin E, Wiggins R, Block KT and Lattanzi R, MR Optimum – A web-based application for signal-to-noise ratio evaluation ; 27th Scientific Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). Montreal (Canada), 11-16 May 2019, p. 4617.
Acknowledgements
This research project is partially supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health (NIH) under Award Number R01 EB024536.










