FireVoxel: Interactive Software for Multi-Modality Analysis of Dynamic Medical Images
Overview
FireVoxel is a medical imaging analysis software that offers an effective set of quantitative tools and has proven to be of particular value for processing dynamic studies in abdominal and genitourinary imaging. Developed and maintained at NYU Langone’s Center for Biomedical Imaging, the software is freely distributed to medical researchers via firevoxel.org.
The FireVoxel package features user-friendly interface and basic image analysis tools such as segmentation, non-uniformity correction, and coregistration, with a recent focus on deformable coregistration and motion correction. The most frequently used FireVoxel methods are dynamic modeling, segmentation, texture analysis and coregistration. The software is often applied in abdominal and genitourinary imaging studies, where it appears to fill a niche due to the lack of alternatives.
As of early 2025, FireVoxel has been downloaded by more than 3,000 users worldwide and has been cited in more than 500 peer-reviewed research articles, nearly 80 percent of which have been MRI studies, with CT and PET studies accounting for about 14 and 7 percent of citations, respectively. The steady increase in publications the software since its first release around 2007 reflects growing interest and continuing relevance for image-based research. The FireVoxel team remains committed to its mission of supporting reliable non-commercial, lab-to-user dissemination of imaging technologies.
Keywords
- Medical Image Analysis
- Dynamic Imaging
- Quantitative Imaging
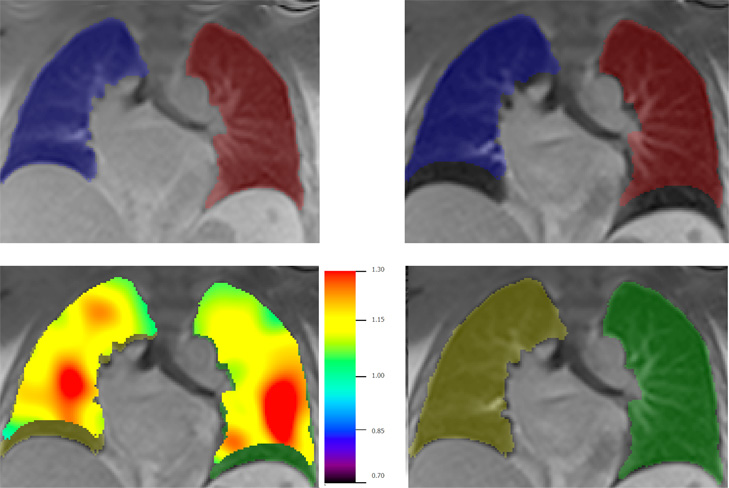
Figure 1. Lung segmentation from dynamic MR images. Segmentation masks are propagated across temporal frames, with left and right lung masks constructed separately. Upper left: initial masks; Upper right: initial masks overlayed on inspiratory frame, showing large mismatch. Lower left: after application of FLEXR, FireVoxel’s elastic deformation algorithm. The color map shows the local Jacobian of estimated transformation. Lower right: resulting segmented right and left lung at inspiratory frame.
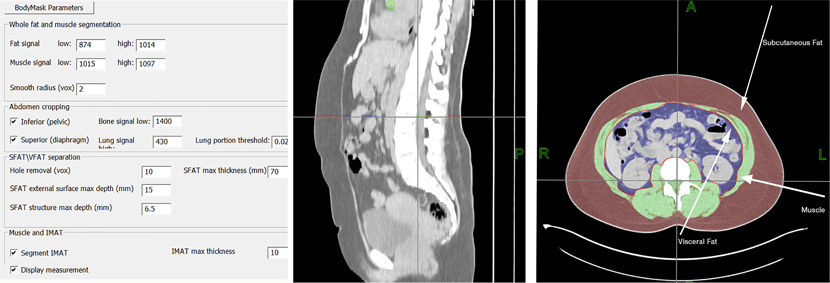
Figure 2. Automatic segmentation of adipose tissue from axial CT images. Left: Approximate ranges of tissue CT attenuation and other parameters used to control the workflow. Middle: The sagittal reformat of the CT through the abdomen. Right: segmentation of visceral (blue) and subcutaneous (red) fat, muscle (green) and intramuscular fat (yellow).
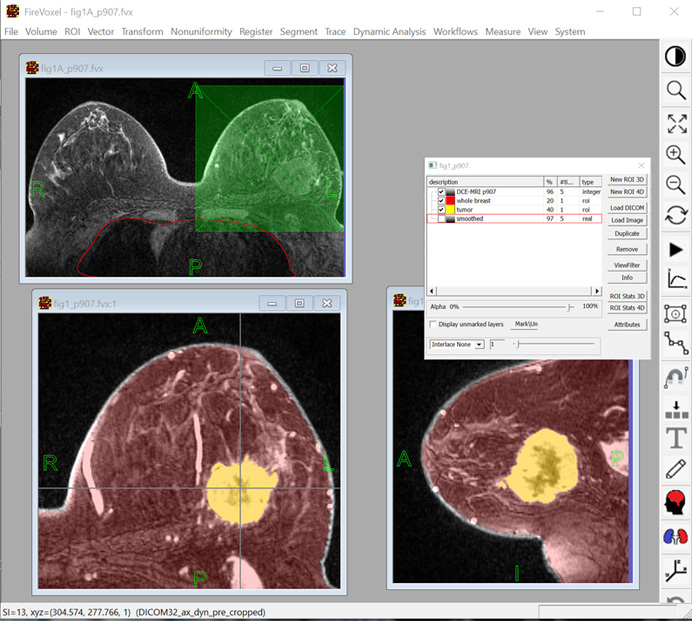
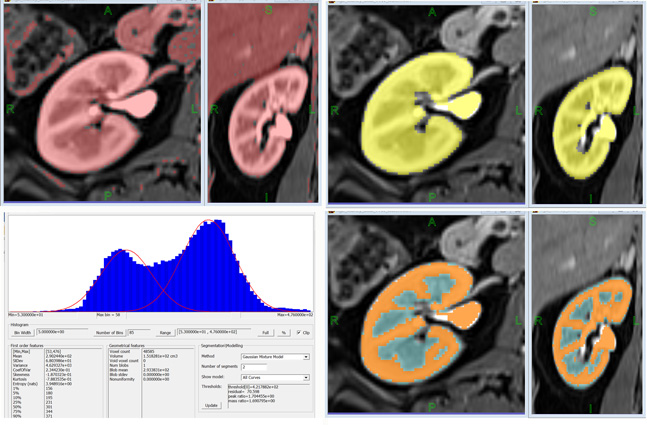
Figure 3. FireVoxel user interface includes of a workspace (gray), a menu of commands and a toolbar. The document at top left contains a T1-weighted fat-suppressed dynamic contrast breast MRI (4D). The document at the bottom is displayed in two linked windows, showing the axial (left) and the sagittal (right) view of the same volume. This document includes two binary masks (ROIs) obtained through image segmentation. Also shown is the Layer Control dialog box used to handle (import, export, transparency, operations, etc.) individual image layers.
Project Team

Henry Rusinek, PhD
Project Lead

Artem Mikheev
Project Lead
External Collaborators
- Louisa Bokacheva, PhD , NYU Langone Health
- Gene S. Kim, PhD, NYU Langone Health
- Samantha L. Heller, MD, PhD, NYU Langone Health
- Lidia Glodzik, Weill Cornell Medical College
- Jeff Lei Zhang, Harvard Medical School
- Ankur M. Doshi, MD, NYU Langone Health
- Ruth Lim, MD, Yale School of Medicine
- Bachir Taouli, MD, Icahn School of Medicine at Mount Sinai
- Anna Chen, MD, NYU Langone Health
- Samir Taneja, MD, NYU Langone Health
- Oded Gonen, PhD, NYU Langone Health
- Uzma Samadani, MD, PhD, University of Minnesota
- Brian Park
- Tracy Butler
- ...and many others
Publications
- Mikheev A, DiMartino JM, Bokacheva L, Rusinek H. FireVoxel: interactive software for multi-modality analysis of dynamic medical images. J Imaging Inform Med, 2025 https://doi.org/10.1007/s10278-025-01404-x.
- Azour L, Rusinek H, Mikheev A, et al. Quantitative Characterization of Respiratory Patterns on Dynamic Higher Temporal Resolution MRI to Stratify Postacute Covid-19 Patients by Cardiopulmonary Symptom Burden. J Magn Reson Imaging. 2024;60(6):2459-2469. doi:10.1002/jmri.29352
- Mikheev A, Nevsky G, Govindan S, Grossman R, Rusinek H. Fully automatic segmentation of the brain from T1-weighted MRI using Bridge Burner algorithm. J Magn Reson Imaging. 2008;27(6):1235-1241. doi:10.1002/jmri.21372
- Chandarana H, Block TK, Ream J, et al. Estimating liver perfusion from free-breathing continuously acquired dynamic gadolinium-ethoxybenzyl-diethylenetriamine pentaacetic acid-enhanced acquisition with compressed sensing reconstruction. Invest Radiol. 2015;50(2):88-94. doi:10.1097/RLI.0000000000000105
- Mikheev A, Logan J, Ding YS, Rusinek H, A novel two-stage iterative vessel tracking algorithm for determining an image-derived input function for PET; J. Nucl. Med., 57, 2016, p. 1905.
Acknowledgements
We acknowledge support from the following NIH grants: U24 EB028980 from NIBIB, U24 NS135568 from NINDS, P41 EB017183 from NIBIB and P30 AG066512 from NIA.









